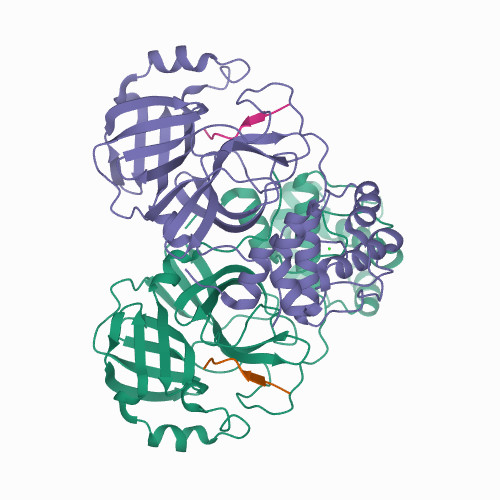
Recognition of Divergent Viral Substrates by the SARS-CoV-2 Main Protease.
Publication Type:
Journal ArticleSource:
ACS Infect Dis (2021)Abstract:
<p>The main protease (M) of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of coronavirus disease (COVID-19), is an ideal target for pharmaceutical inhibition. M is conserved among coronaviruses and distinct from human proteases. Viral replication depends on the cleavage of the viral polyprotein at multiple sites. We present crystal structures of SARS-CoV-2 M bound to two viral substrate peptides. The structures show how M recognizes distinct substrates and how subtle changes in substrate accommodation can drive large changes in catalytic efficiency. One peptide, constituting the junction between viral nonstructural proteins 8 and 9 (nsp8/9), has P1' and P2' residues that are unique among the SARS-CoV-2 M cleavage sites but conserved among homologous junctions in coronaviruses. M cleaves nsp8/9 inefficiently, and amino acid substitutions at P1' or P2' can enhance catalysis. Visualization of M with intact substrates provides new templates for antiviral drug design and suggests that the coronavirus lifecycle selects for finely tuned substrate-dependent catalytic parameters.</p>