Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Publication Type:
Journal ArticleSource:
Proc Natl Acad Sci U S A, Volume 117, Issue 8, p.4109-4116 (2020)Abstract:
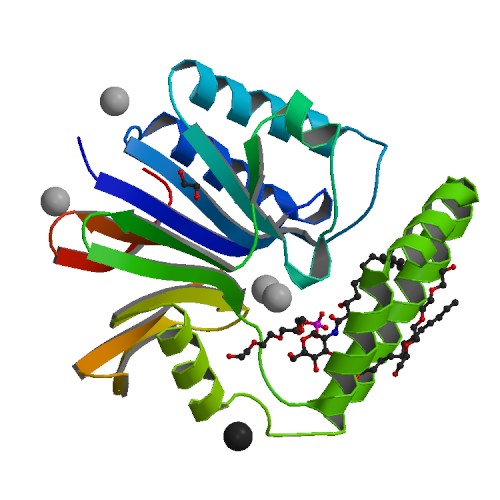
<p>The UDP-2,3-diacylglucosamine pyrophosphate hydrolase LpxH is an essential lipid A biosynthetic enzyme that is conserved in the majority of gram-negative bacteria. It has emerged as an attractive novel antibiotic target due to the recent discovery of an LpxH-targeting sulfonyl piperazine compound (referred to as AZ1) by AstraZeneca. However, the molecular details of AZ1 inhibition have remained unresolved, stymieing further development of this class of antibiotics. Here we report the crystal structure of LpxH in complex with AZ1. We show that AZ1 fits snugly into the -shaped acyl chain-binding chamber of LpxH with its indoline ring situating adjacent to the active site, its sulfonyl group adopting a sharp kink, and its -CF-phenyl substituted piperazine group reaching out to the far side of the LpxH acyl chain-binding chamber. Intriguingly, despite the observation of a single AZ1 conformation in the crystal structure, our solution NMR investigation has revealed the presence of a second ligand conformation invisible in the crystalline state. Together, these distinct ligand conformations delineate a cryptic inhibitor envelope that expands the observed footprint of AZ1 in the LpxH-bound crystal structure and enables the design of AZ1 analogs with enhanced potency in enzymatic assays. These designed compounds display striking improvement in antibiotic activity over AZ1 against wild-type , and coadministration with outer membrane permeability enhancers profoundly sensitizes to designed LpxH inhibitors. Remarkably, none of the sulfonyl piperazine compounds occupies the active site of LpxH, foretelling a straightforward path for rapid optimization of this class of antibiotics.</p>