A Fluorescence Polarization Assay for Macrodomains Facilitates the Identification of Potent Inhibitors of the SARS-CoV-2 Macrodomain.
Publication Type:
Journal ArticleSource:
ACS Chem Biol, Volume 18, Issue 5, p.1200-1207 (2023)Keywords:
Adenosine Diphosphate, Adenosine Diphosphate Ribose, COVID-19, Fluorescence Polarization, Humans, SARS-CoV-2Abstract:
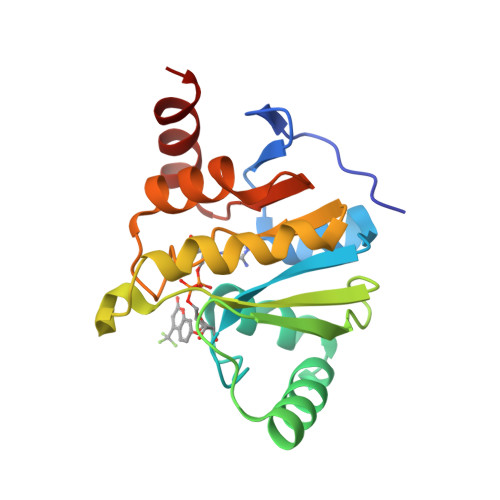
<p>Viral macrodomains, which can bind to and/or hydrolyze adenine diphosphate ribose (ADP-ribose or ADPr) from proteins, have been suggested to counteract host immune response and be viable targets for the development of antiviral drugs. Therefore, developing high-throughput screening (HTS) techniques for macrodomain inhibitors is of great interest. Herein, using a novel tracer , an ADP-ribose compound conjugated with tetramethylrhodamine, we developed a robust fluorescence polarization assay for various viral and human macrodomains including SARS-CoV-2 Macro1, VEEV Macro, CHIKV Macro, human MacroD1, MacroD2, and PARP9 Macro2. Using this assay, we validated (IC 6.4 μM) and (IC 15.2 μM), two literature-reported small-molecule inhibitors of SARS-CoV-2 Macro1. Our data suggest that is highly selective for SARS-CoV-2 Macro1 over other human and viral macrodomains. Furthermore, using this assay, we identified (ADP-ribosylated -nitrophenol, IC 370 nM) and (ADP-ribosylated trifluoromethyl umbelliferone, IC 590 nM) as the most potent SARS-CoV-2 Macro1 binders reported to date. An X-ray crystal structure of SARS-CoV-2 Macro1 in complex with TFMU-ADPr revealed how the TFMU moiety contributes to the binding affinity. Our data demonstrate that this fluorescence polarization assay is a useful addition to the HTS methods for the identification of macrodomain inhibitors.</p>