Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations.
Publication Type:
Journal ArticleSource:
ACS Cent Sci, Volume 7, Issue 3, p.467-475 (2021)Abstract:
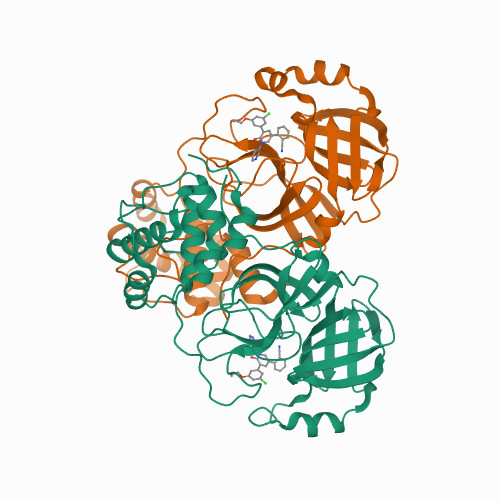
<p>Starting from our previous finding of 14 known drugs as inhibitors of the main protease (M) of SARS-CoV-2, the virus responsible for COVID-19, we have redesigned the weak hit perampanel to yield multiple noncovalent, nonpeptidic inhibitors with ca. 20 nM IC values in a kinetic assay. Free-energy perturbation (FEP) calculations for M-ligand complexes provided valuable guidance on beneficial modifications that rapidly delivered the potent analogues. The design efforts were confirmed and augmented by determination of high-resolution X-ray crystal structures for five analogues bound to M. Results of cell-based antiviral assays further demonstrated the potential of the compounds for treatment of COVID-19. In addition to the possible therapeutic significance, the work clearly demonstrates the power of computational chemistry for drug discovery, especially FEP-guided lead optimization.</p>