A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein
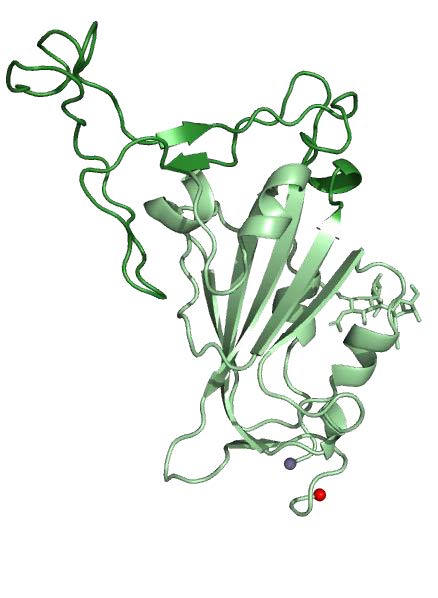
SARS-CoV-2 is a zoonotic virus that has caused a pandemic of severe respiratory disease—COVID-19— within several months of its initial identification. Comparable to the first SARS-CoV, this novel coronavirus’s surface Spike (S) glycoprotein mediates cell entry via the human ACE-2 receptor, and, thus, is the principal target for the development of vaccines and immunotherapeutics. Molecular information on the SARS-CoV-2 S glycoprotein remains limited. Here we report the crystal structure of the SARS-CoV-2 S receptor-binding-domain (RBD) at a the highest resolution to date, of 1.95 Å. We identified a set of SARS-reactive monoclonal antibodies with cross-reactivity to SARS-CoV-2 RBD and other betacoronavirus S glycoproteins. One of these antibodies, CR3022, was previously shown to synergize with antibodies that target the ACE-2 binding site on the SARS-CoV RBD and reduce viral escape capacity. We determined the structure of CR3022, in complex with the SARS-CoV-2 RBD, and defined a broadly reactive epitope that is highly conserved across betacoronaviruses. This epitope is inaccessible in the “closed” prefusion S structure, but is accessible in “open” conformations. This first-ever resolution of a human antibody in complex with SARS-CoV-2 and the broad reactivity of this set of antibodies to a conserved betacoronavirus epitope will allow antigenic assessment of vaccine candidates, and provide a framework for accelerated vaccine, immunotherapeutic and diagnostic strategies against SARS-CoV-2 and related betacoronaviruses.
Work done at NE-CAT on the surface Spike glycoprotein has now been released at the preprint server BioRxiv.
