Research Highlights
(2022) Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants. Emerg Microbes Infect. 11, 548-551
(2022) Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc Natl Acad Sci U S A. 10.1073/pnas.2106379119
(2022) Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL covalent inhibitors.. Eur J Med Chem. 229, 114046

(2022) Crystal structure of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) frameshifting pseudoknot. RNA. 10.1261/rna.078825.121
(2021) Mechanisms of SARS-CoV-2 neutralization by shark variable new antigen receptors elucidated through X-ray crystallography. Nat Commun. 12, 7325

(2021) Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain. Science

(2021) Low-dose in vivo protection and neutralization across SARS-CoV-2 variants by monoclonal antibody combinations. Nat Immunol. 10.1038/s41590-021-01068-z
(2021) A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature. 10.1038/s41586-021-04045-6

(2021) A molecular sensor determines the ubiquitin substrate specificity of SARS-CoV-2 papain-like protease. Cell Rep. 36, 109754
(2021) Structural Basis for SARS-CoV-2 Nucleocapsid Protein Recognition by Single-Domain Antibodies. Front Immunol. 12, 719037

(2021) Recognition of Divergent Viral Substrates by the SARS-CoV-2 Main Protease. ACS Infect Dis. 10.1021/acsinfecdis.1c00237
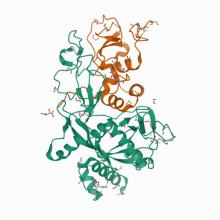
![7N44 Crystal structure of the SARS-CoV-2 (2019-NCoV) main protease in complex with 5-(3-{3-chloro-5-[(5-methyl-1,3-thiazol-4-yl)methoxy]phenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl)pyrimidine-2,4(1H,3H)-dione (compound 13)](https://lilith.nec.aps.anl.gov/sites/default/files/styles/medium/public/7n44_assembly-1.jpeg?itok=V_Uskmkp)
(2021) Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency. ACS Med Chem Lett. 12, 1325-1332
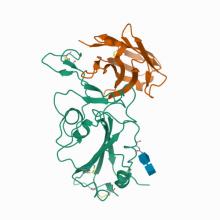
(2021) The development of Nanosota-1 as anti-SARS-CoV-2 nanobody drug candidates. Elife. 10.7554/eLife.64815
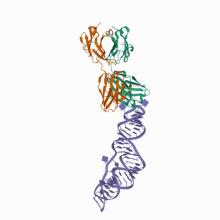
(2021) The SARS-CoV-2 Programmed -1 Ribosomal Frameshifting Element Crystal Structure Solved to 2.09 Å Using Chaperone-Assisted RNA Crystallography.. ACS Chem Biol. 10.1021/acschembio.1c00324
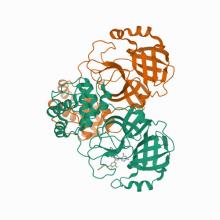
(2021) Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease. Structure. 10.1016/j.str.2021.06.002
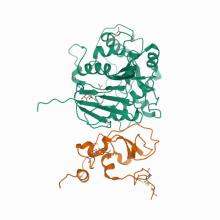
(2021) A metal ion orients SARS-CoV-2 mRNA to ensure accurate 2'-O methylation of its first nucleotide. Nat Commun. 12, 3287
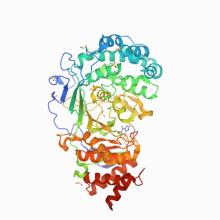
(2021) Targeting SARS-CoV-2 Nsp3 macrodomain structure with insights from human poly(ADP-ribose) glycohydrolase (PARG) structures with inhibitors. Prog Biophys Mol Biol. 10.1016/j.pbiomolbio.2021.02.002
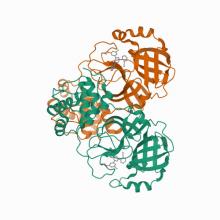
(2021) Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent Sci. 7, 467-475
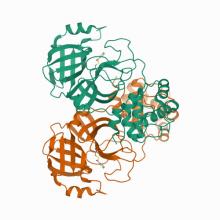
(2021) Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors. Nat Commun. 12, 2016

(2021) Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 10.1016/j.chom.2021.03.005
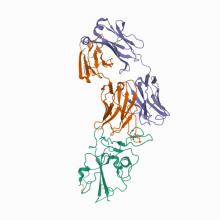
(2021) Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class. Cell Rep. 10.1016/j.celrep.2021.108950

(2021) Inhibition of amyloid formation of the Nucleoprotein of SARS-CoV-2. bioRxiv. 10.1101/2021.03.05.434000
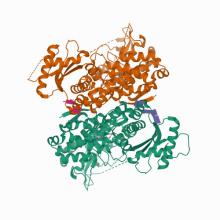
(2021) Nucleic acid binding by SAMHD1 contributes to the antiretroviral activity and is enhanced by the GpsN modification. Nat Commun. 12, 731
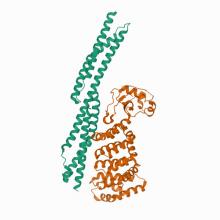
(2021) Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. Structure. 10.1016/j.str.2020.12.013
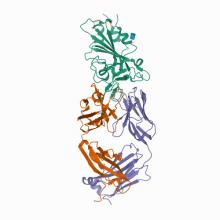
(2021) SARS-CoV-2 evolution in an immunocompromised host reveals shared neutralization escape mechanisms. Cell. 10.1101/2020.11.13.381533

(2020) Biophysical and X-ray structural studies of the (GGGTT)3GGG G-quadruplex in complex with N-methyl mesoporphyrin IX. PLoS One. 15, e0241513
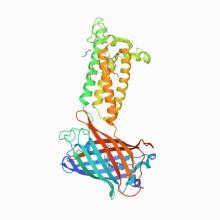
(2020) Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 10.1126/science.abc5667

(2020) Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti-COVID-19 drug design. Sci Adv. 10.1126/sciadv.abd4596
(2020) Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception. Proc Natl Acad Sci U S A. 10.1073/pnas.1920444117

(2020) A defined structural unit enables de novo design of small-molecule-binding proteins. Science. 369, 1227-1233
(2020) Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 10.1002/pro.3909
(2020) A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. bioRxiv. 10.1101/2020.03.15.992883