Research Highlights
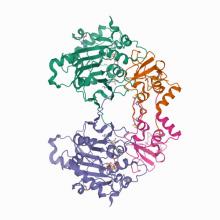
(2020) Architecture and self-assembly of the SARS-CoV-2 nucleocapsid protein. Protein Sci. 10.1002/pro.3909
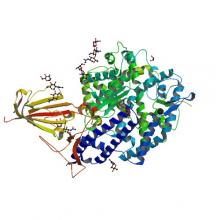
(2020) A Cryptic Site of Vulnerability on the Receptor Binding Domain of the SARS-CoV-2 Spike Glycoprotein. bioRxiv. 10.1101/2020.03.15.992883
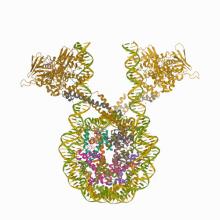
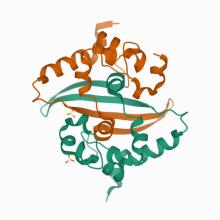
(2020) Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate. Mol Cell. 10.1016/j.molcel.2020.04.019
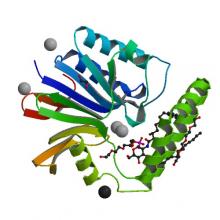
(2020) Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics. Proc Natl Acad Sci U S A. 117, 4109-4116
(2020) The human dimension in contemporary biological research. Nat Struct Mol Biol. 27, 107-108
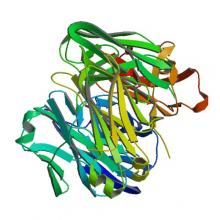
(2020) Mechanism of differential Zika and dengue virus neutralization by a public antibody lineage targeting the DIII lateral ridge. J Exp Med. 10.1084/jem.20191792
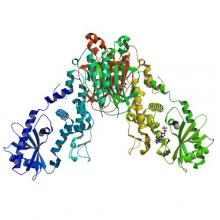
(2019) HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity. Mol Cell. 10.1016/j.molcel.2019.12.009
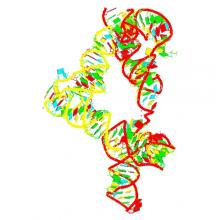
(2019) Structural basis for tRNA decoding and aminoacylation sensing by T-box riboregulators. Nat Struct Mol Biol. 26, 1106-1113

(2019) Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis. Proc Natl Acad Sci U S A. 116, 17825-17830
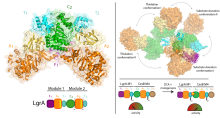
(2019) Structures of a dimodular nonribosomal peptide synthetase reveal conformational flexibility. Science. 10.1126/science.aaw4388
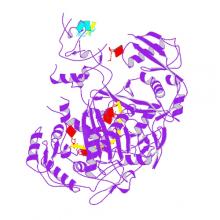
(2019) Multidomain Convergence of Argonaute during RISC Assembly Correlates with the Formation of Internal Water Clusters. Mol Cell. 75, 725-740.e6
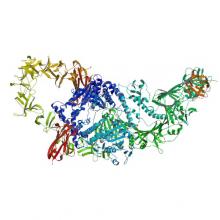
(2019) Structure of the full-length Clostridium difficile toxin B. Nat Struct Mol Biol. 10.1038/s41594-019-0268-0
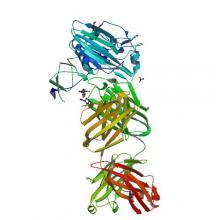
(2019) Antibodies to a Conserved Influenza Head Interface Epitope Protect by an IgG Subtype-Dependent Mechanism. Cell. 177, 1124-1135.e16

(2019) Visualization of clustered protocadherin neuronal self-recognition complexes. Nature. 569, 280-283

(2019) Bacterial cGAS-like enzymes synthesize diverse nucleotide signals. Nature. 10.1038/s41586-019-0953-5
![Structure of VACV Poxin in post-reactive state with Gp[2'-5']Ap[3'] Structure of VACV Poxin in post-reactive state with Gp[2'-5']Ap[3']](https://lilith.nec.aps.anl.gov/sites/default/files/styles/medium/public/6ea9.pdb1-500_0.jpg?itok=g_h_U9hi)
(2019) Viral and metazoan poxins are cGAMP-specific nucleases that restrict cGAS-STING signalling. Nature. 566, 259-263
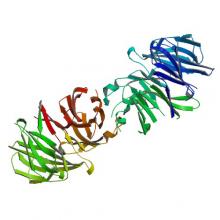
(2019) Crystal structure of the WD40 domain dimer of LRRK2. Proc Natl Acad Sci U S A. 10.1073/pnas.1817889116
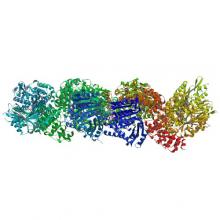
(2018) Structural basis of tubulin recruitment and assembly by microtubule polymerases with Tumor Overexpressed Gene (TOG) domain arrays. Elife. 10.7554/eLife.38922

(2018) Redox-dependent rearrangements of the NiFeS cluster of carbon monoxide dehydrogenase. Elife. 10.7554/eLife.39451
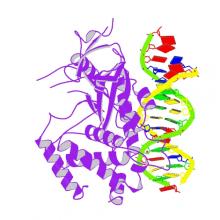
(2018) Structure of the Human cGAS-DNA Complex Reveals Enhanced Control of Immune Surveillance. Cell. 174, 300-311.e11
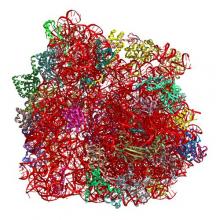
(2018) Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site. Mol Cell. 70, 83-94.e7
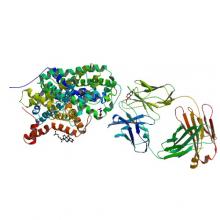
(2018) Structural basis for recognition of diverse antidepressants by the human serotonin transporter. Nat Struct Mol Biol. 10.1038/s41594-018-0026-8
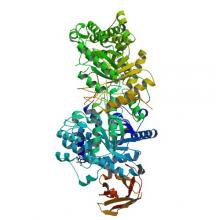
(2018) Structures of β-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.. Nature. 10.1038/nature25010
(2017) Non-specific activities of the major herbicide-resistance gene BAR. Nat Plants. 3, 937-945
(2017) Structural basis for methylphosphonate biosynthesis. Science. 358, 1336-1339
(2017) How type II CRISPR-Cas establish immunity through Cas1-Cas2-mediated spacer integration. Nature. 550, 137-141

(2017) The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature. 10.1038/nature23484
(2017) Inhibition Mechanism of an Anti-CRISPR Suppressor AcrIIA4 Targeting SpyCas9. Mol Cell. 10.1016/j.molcel.2017.05.024
(2017) RNA polymerase motions during promoter melting. Science. 356, 863-866

(2017) Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode. Acta Crystallogr D Struct Biol. 73, 294-315
(2017) Crystal Structure of the Receptor-Binding Domain of Botulinum Neurotoxin Type HA, Also Known as Type FA or H. Toxins (Basel). 10.3390/toxins9030093